Note
This tutorial was generated from an IPython notebook that can be downloaded here.
Create 3D region masks¶
In this tutorial we will show how to create 3D boolean masks for
arbitrary latitude and longitude grids. It uses the same algorithm to
determine if a gridpoint is in a region as for the 2D mask. However, it
returns a xarray.Dataset with shape region x lat x lon,
gridpoints that do not fall in a region are False, the gridpoints
that fall in a region are True.
Import regionmask and check the version:
import regionmask
regionmask.__version__
'0.5.0+dev'
Load xarray and numpy:
import xarray as xr
import numpy as np
Creating a mask¶
We define a lon/ lat grid with a 1° grid spacing, where the points define the center of the grid.
lon = np.arange(-179.5, 180)
lat = np.arange(-89.5, 90)
We create a mask with the SREX regions (Seneviratne et al., 2012).
regionmask.defined_regions.srex
<regionmask.Regions>
Name: SREX
Source: Seneviratne et al., 2012 (https://www.ipcc.ch/site/assets/uploads/2...
Regions:
1 ALA Alaska/N.W. Canada
2 CGI Canada/Greenl./Icel.
3 WNA W. North America
4 CNA C. North America
5 ENA E. North America
.. ... ...
22 EAS E. Asia
23 SAS S. Asia
24 SEA S.E. Asia
25 NAU N. Australia
26 SAU S. Australia/New Zealand
[26 regions]
The function mask_3D determines which gripoints lie within the
polygon making up the each region:
mask = regionmask.defined_regions.srex.mask_3D(lon, lat)
mask
- region: 26
- lat: 180
- lon: 360
- False False False False False False ... False False False False False
array([[[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], ..., [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]]]) - lon(lon)float64-179.5 -178.5 ... 178.5 179.5
array([-179.5, -178.5, -177.5, ..., 177.5, 178.5, 179.5])
- lat(lat)float64-89.5 -88.5 -87.5 ... 88.5 89.5
array([-89.5, -88.5, -87.5, -86.5, -85.5, -84.5, -83.5, -82.5, -81.5, -80.5, -79.5, -78.5, -77.5, -76.5, -75.5, -74.5, -73.5, -72.5, -71.5, -70.5, -69.5, -68.5, -67.5, -66.5, -65.5, -64.5, -63.5, -62.5, -61.5, -60.5, -59.5, -58.5, -57.5, -56.5, -55.5, -54.5, -53.5, -52.5, -51.5, -50.5, -49.5, -48.5, -47.5, -46.5, -45.5, -44.5, -43.5, -42.5, -41.5, -40.5, -39.5, -38.5, -37.5, -36.5, -35.5, -34.5, -33.5, -32.5, -31.5, -30.5, -29.5, -28.5, -27.5, -26.5, -25.5, -24.5, -23.5, -22.5, -21.5, -20.5, -19.5, -18.5, -17.5, -16.5, -15.5, -14.5, -13.5, -12.5, -11.5, -10.5, -9.5, -8.5, -7.5, -6.5, -5.5, -4.5, -3.5, -2.5, -1.5, -0.5, 0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 10.5, 11.5, 12.5, 13.5, 14.5, 15.5, 16.5, 17.5, 18.5, 19.5, 20.5, 21.5, 22.5, 23.5, 24.5, 25.5, 26.5, 27.5, 28.5, 29.5, 30.5, 31.5, 32.5, 33.5, 34.5, 35.5, 36.5, 37.5, 38.5, 39.5, 40.5, 41.5, 42.5, 43.5, 44.5, 45.5, 46.5, 47.5, 48.5, 49.5, 50.5, 51.5, 52.5, 53.5, 54.5, 55.5, 56.5, 57.5, 58.5, 59.5, 60.5, 61.5, 62.5, 63.5, 64.5, 65.5, 66.5, 67.5, 68.5, 69.5, 70.5, 71.5, 72.5, 73.5, 74.5, 75.5, 76.5, 77.5, 78.5, 79.5, 80.5, 81.5, 82.5, 83.5, 84.5, 85.5, 86.5, 87.5, 88.5, 89.5]) - region(region)int641 2 3 4 5 6 7 ... 21 22 23 24 25 26
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26]) - abbrev(region)<U3'ALA' 'CGI' 'WNA' ... 'NAU' 'SAU'
array(['ALA', 'CGI', 'WNA', 'CNA', 'ENA', 'CAM', 'AMZ', 'NEB', 'WSA', 'SSA', 'NEU', 'CEU', 'MED', 'SAH', 'WAF', 'EAF', 'SAF', 'NAS', 'WAS', 'CAS', 'TIB', 'EAS', 'SAS', 'SEA', 'NAU', 'SAU'], dtype='<U3') - name(region)<U24'Alaska/N.W. Canada' ... 'S. Australia/New Zealand'
array(['Alaska/N.W. Canada', 'Canada/Greenl./Icel.', 'W. North America', 'C. North America', 'E. North America', 'Central America/Mexico', 'Amazon', 'N.E. Brazil', 'Coast South America', 'S.E. South America', 'N. Europe', 'C. Europe', 'S. Europe/Mediterranean', 'Sahara', 'W. Africa', 'E. Africa', 'S. Africa', 'N. Asia', 'W. Asia', 'C. Asia', 'Tibetan Plateau', 'E. Asia', 'S. Asia', 'S.E. Asia', 'N. Australia', 'S. Australia/New Zealand'], dtype='<U24')
As mentioned, mask is a boolean xarray.Dataset with shape
region x lat x lon. It contains region as dimension coordinate
and abbrev and name as non-dimension coordinates (see the xarray
docs for the details on the
terminology).
The four first layers look as follows:
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
from matplotlib import colors as mplc
cmap1 = mplc.ListedColormap(["none", "#9ecae1"])
fg = mask.isel(region=slice(4)).plot(
subplot_kws=dict(projection=ccrs.PlateCarree()),
col="region",
col_wrap=2,
transform=ccrs.PlateCarree(),
add_colorbar=False,
aspect=1.5,
cmap=cmap1,
)
for ax in fg.axes.flatten():
ax.coastlines()
fg.fig.subplots_adjust(hspace=0, wspace=0.1);
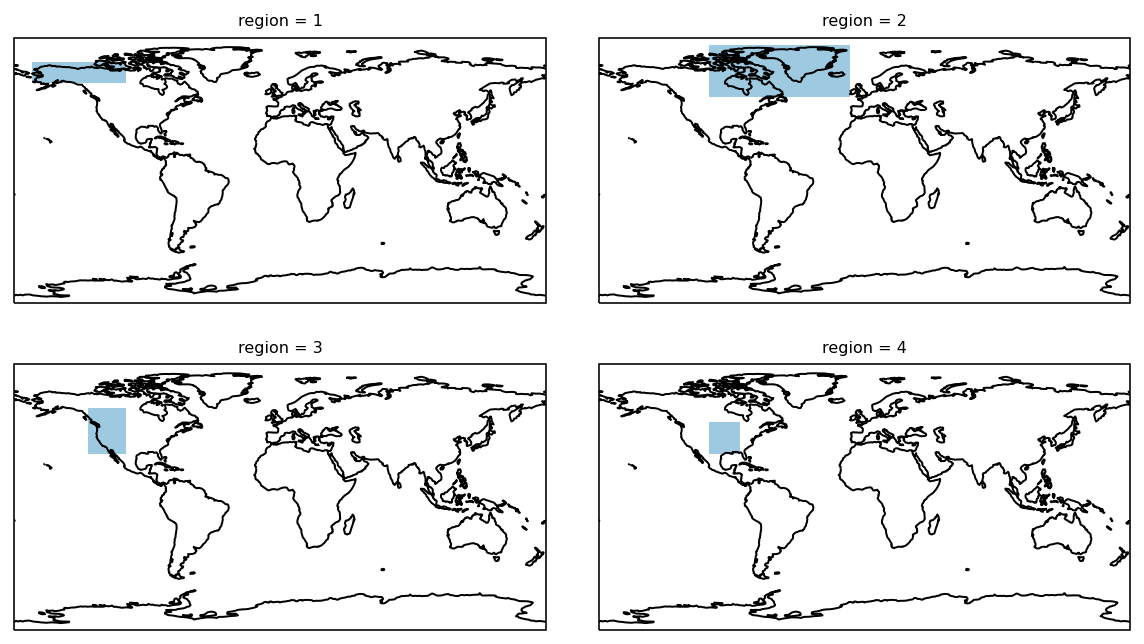
Working with a 3D mask¶
masks can be used to select data in a certain region and to calculate regional averages - let’s illustrate this with a ‘real’ dataset:
airtemps = xr.tutorial.load_dataset("air_temperature")
The example data is a temperature field over North America. Let’s plot the first time step:
# choose a good projection for regional maps
proj = ccrs.LambertConformal(central_longitude=-100)
ax = plt.subplot(111, projection=proj)
airtemps.isel(time=1).air.plot.pcolormesh(ax=ax, transform=ccrs.PlateCarree())
ax.coastlines();
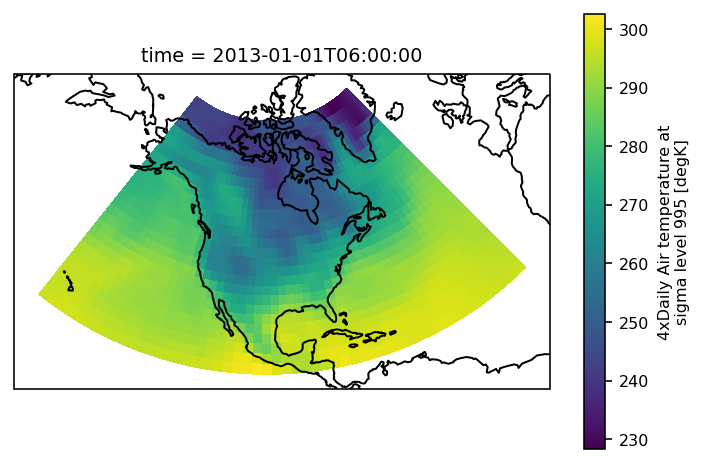
An xarray object can be passed to the mask_3D function:
mask_full = regionmask.defined_regions.srex.mask_3D(airtemps)
mask_full
- region: 26
- lat: 25
- lon: 53
- False False False False False False ... False False False False False
array([[[False, False, False, ..., False, False, False], [ True, True, True, ..., False, False, False], [ True, True, True, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., True, True, True], [False, False, False, ..., True, True, True], [False, False, False, ..., True, True, True], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], ..., [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]]]) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32) - lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - region(region)int641 2 3 4 5 6 7 ... 21 22 23 24 25 26
array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26]) - abbrev(region)<U3'ALA' 'CGI' 'WNA' ... 'NAU' 'SAU'
array(['ALA', 'CGI', 'WNA', 'CNA', 'ENA', 'CAM', 'AMZ', 'NEB', 'WSA', 'SSA', 'NEU', 'CEU', 'MED', 'SAH', 'WAF', 'EAF', 'SAF', 'NAS', 'WAS', 'CAS', 'TIB', 'EAS', 'SAS', 'SEA', 'NAU', 'SAU'], dtype='<U3') - name(region)<U24'Alaska/N.W. Canada' ... 'S. Australia/New Zealand'
array(['Alaska/N.W. Canada', 'Canada/Greenl./Icel.', 'W. North America', 'C. North America', 'E. North America', 'Central America/Mexico', 'Amazon', 'N.E. Brazil', 'Coast South America', 'S.E. South America', 'N. Europe', 'C. Europe', 'S. Europe/Mediterranean', 'Sahara', 'W. Africa', 'E. Africa', 'S. Africa', 'N. Asia', 'W. Asia', 'C. Asia', 'Tibetan Plateau', 'E. Asia', 'S. Asia', 'S.E. Asia', 'N. Australia', 'S. Australia/New Zealand'], dtype='<U24')
Per default this creates a mask containing one layer for each
individual region - even if no gridpoint is contained in this region
(all points are False). As the example data only has values over
Northern America most layers will only contain False. To only obtain
those layers containing (at least) one gridpoint specify drop=True:
mask_drop = regionmask.defined_regions.srex.mask_3D(airtemps, drop=True)
mask_drop
- region: 6
- lat: 25
- lon: 53
- False False False False False False ... False False False False False
array([[[False, False, False, ..., False, False, False], [ True, True, True, ..., False, False, False], [ True, True, True, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., True, True, True], [False, False, False, ..., True, True, True], [False, False, False, ..., True, True, True], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]]]) - lon(lon)float32200.0 202.5 205.0 ... 327.5 330.0
array([200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. ], dtype=float32) - lat(lat)float3275.0 72.5 70.0 ... 20.0 17.5 15.0
array([75. , 72.5, 70. , 67.5, 65. , 62.5, 60. , 57.5, 55. , 52.5, 50. , 47.5, 45. , 42.5, 40. , 37.5, 35. , 32.5, 30. , 27.5, 25. , 22.5, 20. , 17.5, 15. ], dtype=float32) - region(region)int641 2 3 4 5 6
array([1, 2, 3, 4, 5, 6])
- abbrev(region)<U3'ALA' 'CGI' 'WNA' 'CNA' 'ENA' 'CAM'
array(['ALA', 'CGI', 'WNA', 'CNA', 'ENA', 'CAM'], dtype='<U3')
- name(region)<U22'Alaska/N.W. Canada' ... 'Central America/Mexico'
array(['Alaska/N.W. Canada', 'Canada/Greenl./Icel.', 'W. North America', 'C. North America', 'E. North America', 'Central America/Mexico'], dtype='<U22')
Note how there are only 6 layers.
Calculate weighted regional averages¶
Using the 3-dimensional mask it is possible to calculate weighted
averages of all regions in one go, using the weighted method
(requires xarray 0.15.1 or later). As proxy of the grid cell area we use
cos(lat):
weights = np.cos(np.deg2rad(airtemps.lat))
ts_airtemps_regional = airtemps.weighted(mask_drop * weights).mean(dim=("lat", "lon"))
ts_airtemps_regional
- region: 6
- time: 2920
- time(time)datetime64[ns]2013-01-01 ... 2014-12-31T18:00:00
- standard_name :
- time
- long_name :
- Time
array(['2013-01-01T00:00:00.000000000', '2013-01-01T06:00:00.000000000', '2013-01-01T12:00:00.000000000', ..., '2014-12-31T06:00:00.000000000', '2014-12-31T12:00:00.000000000', '2014-12-31T18:00:00.000000000'], dtype='datetime64[ns]') - region(region)int641 2 3 4 5 6
array([1, 2, 3, 4, 5, 6])
- abbrev(region)<U3'ALA' 'CGI' 'WNA' 'CNA' 'ENA' 'CAM'
array(['ALA', 'CGI', 'WNA', 'CNA', 'ENA', 'CAM'], dtype='<U3')
- name(region)<U22'Alaska/N.W. Canada' ... 'Central America/Mexico'
array(['Alaska/N.W. Canada', 'Canada/Greenl./Icel.', 'W. North America', 'C. North America', 'E. North America', 'Central America/Mexico'], dtype='<U22')
- air(time, region)float32255.46873 255.63712 ... 295.03595
array([[255.46873, 255.63712, 272.55185, 271.25925, 279.89474, 295.0278 ], [255.18092, 254.97868, 270.3979 , 269.2749 , 279.28098, 293.58374], [255.03746, 255.30022, 269.289 , 267.79205, 279.26285, 292.88605], ..., [260.47092, 253.83435, 268.54593, 261.13162, 275.96317, 293.93817], [258.43375, 254.55461, 269.3965 , 262.11487, 275.55936, 293.37787], [256.5124 , 254.48091, 270.21487, 266.05923, 276.97205, 295.03595]], dtype=float32)
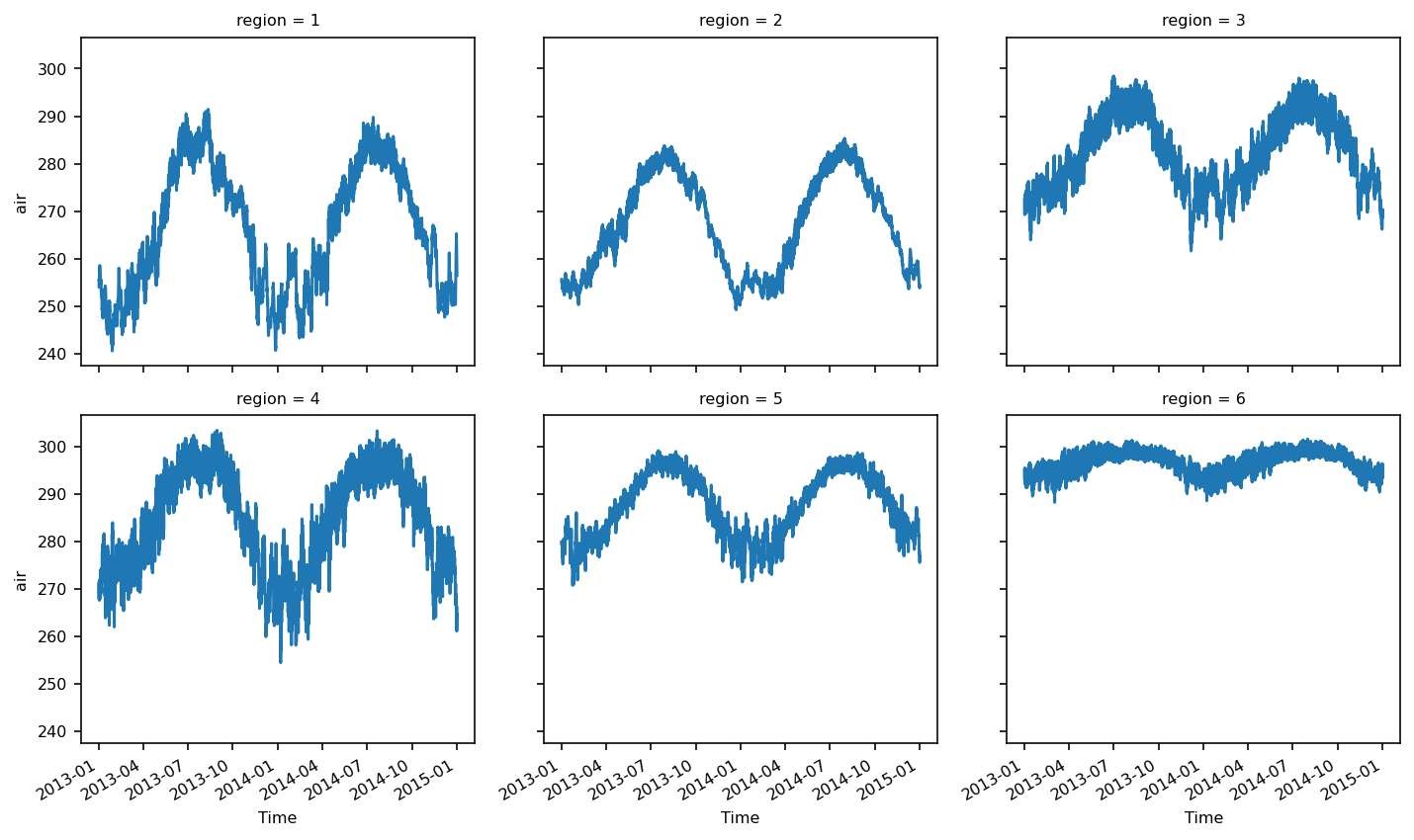
The regionally-averaged time series can be plotted:
ts_airtemps_regional.air.plot(col="region", col_wrap=3);
Restrict the mask to land points¶
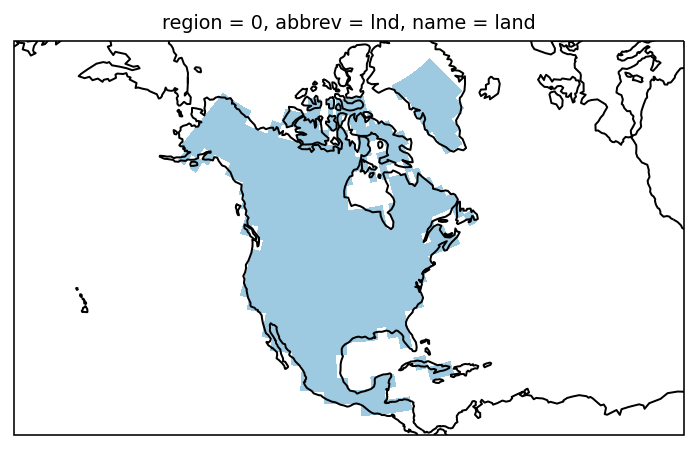
Combining the mask of the regions with a land-sea mask we can create a
land-only mask. We use the natural_earth.land_110 regions to create
a land-sea mask:
land_110 = regionmask.defined_regions.natural_earth.land_110
land_sea_mask = land_110.mask_3D(airtemps)
and plot it
proj = ccrs.LambertConformal(central_longitude=-100)
ax = plt.subplot(111, projection=proj)
land_sea_mask.squeeze().plot.pcolormesh(
ax=ax, transform=ccrs.PlateCarree(), cmap=cmap1, add_colorbar=False
)
ax.coastlines();
To create the combined mask we multiply the two:
mask_lsm = mask_drop * land_sea_mask.squeeze(drop=True)
Note the squeeze(drop=True). This is required to remove the regions
dimension from land_sea_mask.
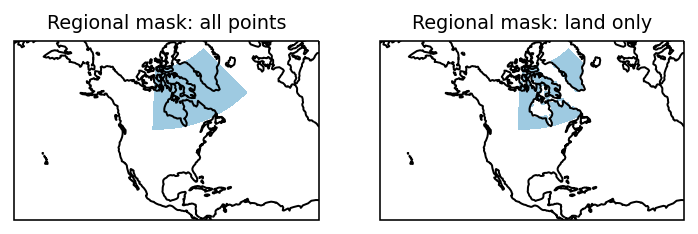
Finally we compare the original mask with the one restricted to land points:
f, axes = plt.subplots(1, 2, subplot_kw=dict(projection=proj))
ax = axes[0]
mask_drop.sel(region=2).plot(
ax=ax, transform=ccrs.PlateCarree(), add_colorbar=False, cmap=cmap1
)
ax.coastlines()
ax.set_title("Regional mask: all points")
ax = axes[1]
mask_lsm.sel(region=2).plot(
ax=ax, transform=ccrs.PlateCarree(), add_colorbar=False, cmap=cmap1
)
ax.coastlines()
ax.set_title("Regional mask: land only");
References¶
Special Report on Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation (SREX, Seneviratne et al., 2012: https://www.ipcc.ch/site/assets/uploads/2018/03/SREX-Ch3-Supplement_FINAL-1.pdf)